McSplit Maximum Common Subgraph Algorithm
Introduction
Maximum common subgraph (MCS) isomorphism algorithms play an important role in chemoinformatics by providing an effective mechanism for the alignment of pairs of chemical structures. It has many chemoinformatics applications, such as, identifying chemical reaction sites (reaction atom mapping), bioactivity prediction of compounds, and exploring structure-activity relationships. Except chemoinformatics, MCS algorithms are used in many other disciplines, and many different MCS algorithms have been developed, but due to the complexity of MCS problems(NP-hard), there is no an effective MCS algorithm until now.
Maximum common subgraph
Before introduce the McSplit algorithm, I must describe some basic definitions.
Induced subgraph of a graph is another graph, formed from a subset of the vertices of the graph and all of the edges (from the original graph) connecting pairs of vertices in that subset. It means for each pair of vertices, if it have an edge in original graph, this edge must be also the induced graph.
Edge-induced subgraph is a set of edges taken from the original graph, in which vertices connected to the edges are included.
A subgraph is a common subgraph of graphs G and H if it is isomorphic to the subgraphs G' and H' of G and H respectively.
There are types of Maximum Common Subgraph. The Maximum Common Induced Subgraph (MCIS) is the largest (in terms of the number of vertices) induced subgraph common to G and H, whereas the Maximum Common Edge Subgraph (MCES) is to find a graph with as many edges as possible that is isomorphic to a subgraph of G and to a subgraph of H simultaneously. These subgraphs are not required to be induced; that is, there can be edges present in either input graph that are not present in the subgraph. There are two types of MCES: the connected MCES (cMCES) and the disconnected MCES (dMCES). A cMCES is one in which each constituent vertex is connected by at least one path in the graph, i.e., the MCES consists of a single, connected subgraph; in a dMCES, conversely, this condition does not hold and the MCES can hence contain two or more subgraphs.
McSplit algorithm
McSplit is one of the state of the art MCS algorithms. It is presented in 2017 by McCreesh et al. It is more than an order of magnitude faster than the previous state of the art for unlabelled and undirected MCS instances. It is also extended for labelled graphs. It is a vertex labelling and partitioning based branch and bound algorithm. The key idea is illustrated as below figure.
In the initialization stage, we partition the input graphs vertices into different classes according the vertex label. Then selecting a vertex matched pair into the matching set, and partition other vertices into new classed by the vertex label and adjacence. For example, in the above figure, after selecting the first pair, the vertices are split into two classes, purple color vertices are adjacent to the selected pair, while green color vertices are not adjacent to the selected pair. Then select the next pair vetices from the two graphs with the same class color, and split vertices into new classes.
The algorithm backtracks when the incumbent (the largest mapping found so far) is at least as large as a calculated bound given M and the current labelling. In the selecting next matched pair procedure, McSplit adopts a heuristic that selects node pairs with the largest degree as its policy. Below is the pesudo-code of the McSplit algorithm.
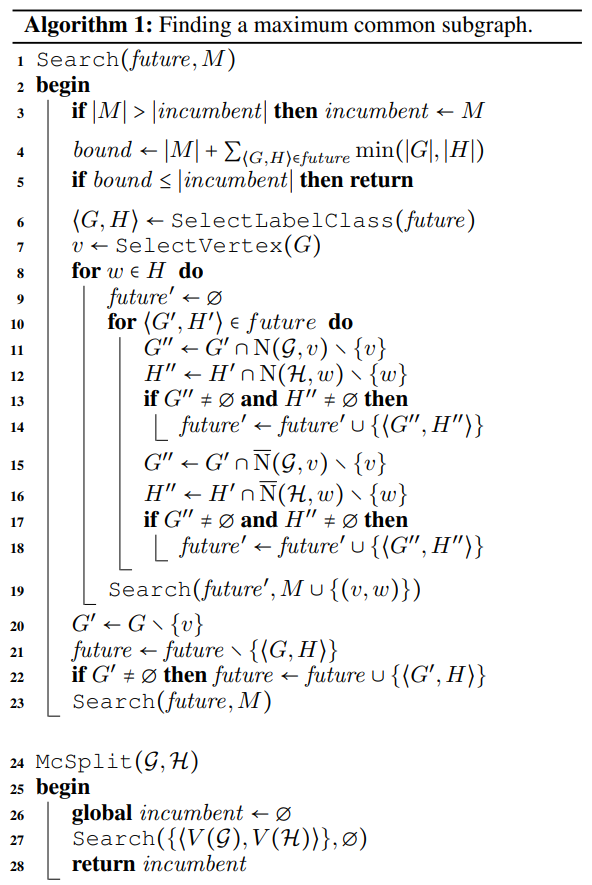
The original McSplit algorithm is used for MCIS, not MCES. However, the MCES better describes the notion of chemical similarity, and finding the MCES is computationally simpler than finding the MCIS. This is because in the former case, one must take into account both vertex and edge labels, rather than just vertex labels as in the MCIS, with the result that the modular product will contain fewer vertices and edges. The problem of identifying the MCIS can be converted to that of identifying the MCES using a theorem due to Whitney and by Nicholson. If want to find the MCES, just need to convert the original graph to line graph, and then perform the algorithm on the line graph. It should be noted that there is one exception to Whitney’s theorem. This is the so-called \(\Delta\) - Y exchange where two different graphs lead to isomorphic edge-graphs and hence to erroneous results for the MCES when translating back from line graphs to the original graphs: however, such results are easy to identify and prune, hence ensuring the identification of the correct MCES.
Conclusion
The MCS is very helpful in chemoinformatics, however, MCS computation is NP-hard, and state-of-the-art exact MCS solvers do not have worst-case time complexity guarantee and cannot handle large graphs in practice. Designing learning based models to find the MCS between two graphs in an approximate yet accurate way while utilizing as few labeled MCS instances as possible remains to be a challenging task. This post introduce the one of the fastest MCS algorithms, McSplit.